Reading time: 4 minutes
Aya Elmeligy
Tumor heterogeneity and lung cancer progression
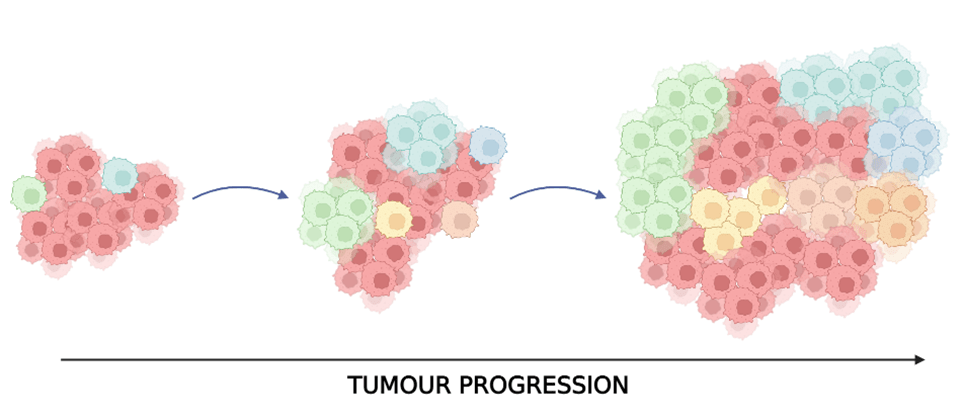
Intratumor heterogeneity is a key concept in the evolution and progression of many tumors, including lung cancers. As a tumor grows, its DNA continually develops mutations to overcome the effects of therapies. The cells that acquire these mutations are known as a subclone. Considering this concept in evolutionary terms, only certain subclones will selectively survive while others are susceptible to treatment.
The same idea applies to lung cancer, the 3rd most common cancer in the UK, with around 48,500 people diagnosed yearly. Most of these cases (80-85%) are a subtype called non-small cell lung cancer. The secrets and pathways through which lung cancer evolves and metastasizes (spreading of the tumor cells to other parts of the body) are essential for developing better prognostic tools in the clinic.
TRACERx Study
TracerX is a large study spanning over 19 centers within the UK, aiming to collect genomic and clinical data from over 800 patients with NSCLC to develop more effective lung cancer treatments. This comprehensive study followed patients throughout their treatment regimen to understand the behavior of cells within the same tumor and across patients. This flagship study found that tumor diversity and heterogeneity were positively correlated with patient relapse. A recent paper compared primary and metastatic tumor samples of patients whose tumors had spread with patients whose cancer had not metastasized (1). This research tracked the evolution of early-stage non-small cell lung cancer over time by following patient experiences through the healthcare system. Primary tumor samples were taken from the 421 recruited patients, and 96 samples were sequenced. After a median follow-up of 4.66 years, 142 out of 421 patients had a recurrence. From these, ‘metastatic’ samples were successfully taken from 48 patients. Comparisons of matched primary and metastatic tumor samples could be made from the collected data. Researchers termed the most recently shared clone between a primary tumor and metastasis as ‘seeding clones.’ Metastatic seeding clones drove evolution as they appeared fitter and occupied more of the tumor. Data showed that 75% of metastases diverge in the later stages of cancer. Those diverging early were positively correlated with smoking habits, which allows cancer cells to adapt to their environment. Furthermore, platinum chemotherapy was associated with tumor heterogeneity and cancer cell evolution.
An additional paper paired the use of both genomic and transcriptomic analysis of non-small cell lung cancer (NSCLC) samples to highlight any sources of variation to characterize the phenotype of the metastatic seeding region within primary tumors (2). Research showed that disruptive allele-specific expression occurred due to chromosomal instability in most tumor samples. Allele-specific expression is associated with DNA methylation and inactivating mutations in epigenetic modifier genes SETD2 (histone methyltransferase gene) and KDM5C (lysine methylase gene), and KMT2B (lysine methyltransferase gene). Epigenetic modifier genes are those linked to alterations within the DNA post-transcriptionally. Further data provide insights into how the loss of SETD2 leads to increased oncogenic transcriptional output. Additionally, KDM5C can regulate transcription through H3K4 demethylation. This study shows that transcriptional variation plays a key role in NSCLC evolution, and therefore the sources of diversity discovered in this study provide insight into how tumors progress.
Future prospects
Predicting the metastatic journey of a tumor through an understanding of this evolution is a vital research area to help develop more personalized treatment methods for use in the clinic. The TracerX study has successfully acquired multiple biopsies throughout a patient’s treatment, allowing large amounts of data to be collected on how tumors evolve in response to treatment. To improve upon these studies, less invasive approaches, such as those that seek to acquire and analyze circulating tumor DNA to follow the rise of seeding clones, are necessary (1).
The transcriptomic variation found within lung tumors cannot fully show the extent of tumor evolution since factors such as alternative splicing are not included. Furthermore, only a small portion of RNA editing was considered, meaning the true level of variation within a tumor sample was likely underrepresented by the data collected. However, the data does show that transcriptional variation is a key contributor to the progression and evolution of NSCLC. This study has paved the way for future research to delve deeper into this variation and highlight the specific consequences of tumor evolution (2).
Edited by Alex Woodell
References
1. Al Bakir, M., Huebner, A., Martínez-Ruiz, C., Grigoriadis, K., Watkins, T.B.K., Pich, O., Moore, D.A., Veeriah, S., Ward, S., Laycock, J., Johnson, D., Rowan, A., Razaq, M., Akther, M., Naceur-Lombardelli, C., Prymas, P., Toncheva, A., Hessey, S., Dietzen, M. and Colliver, E. (2023). The evolution of non-small cell lung cancer metastases in TRACERx. Nature, 616(7957), pp.534–542.
2. Martínez-Ruiz, C., Black, J.R.M., Puttick, C., Hill, M.S., Demeulemeester, J., Larose Cadieux, E., Thol, K., Jones, T.P., Veeriah, S., Naceur-Lombardelli, C., Toncheva, A., Prymas, P., Rowan, A., Ward, S., Cubitt, L., Athanasopoulou, F., Pich, O., Karasaki, T., Moore, D.A. and Salgado, R. (2023). Genomic–transcriptomic evolution in lung cancer and metastasis. Nature, 616(7957), pp.543–552.
3. Image made with Biorender.com
Leave a comment